Welcome to UTM Biology Seminar Series 2023-2024
When: Fridays, 12-1pm
Where: DV2072 (Fall)
IB335 (Winter)
Every Friday during the academic year, the Department of Biology hosts an exciting seminar given by a guest speaker. Topics cover every aspect of biology, from whole organisms to molecular biology,
Everyone is welcome to attend!
https://utoronto.zoom.us/j/84984673393
Kelly Murray-Stoker, PhD student, McCauley Lab
Title: Forces structuring caddisfly communities: Biogeographical to anthropogenic effects
Abstract: Species assemblages result from complex interactions of a range of factors: abiotic or biotic; anthropogenic or not; short-term or long-term; local or regional. I am interested in how environmental factors at different scales interact to influence community diversity of Trichoptera (caddisflies), an aquatic insect order, in the Nearctic bioregion. My thesis focuses on (1.) how taxonomic and functional diversity of stream caddisfly communities vary across geographic regions and environmental gradients, (2.) whether environmental determinants of diversity depend upon species traits, and (3.) whether there is variation in how caddisfly communities respond to urbanization due to geographical context. To address the diversity of caddisfly communities across different ecoregions, I compiled data from the U.S. Environmental Protection Agency’s National Rivers and Streams Assessment; preliminary results point to the importance of ecoregion to community structure and taxonomic and functional trait richness. In characterizing patterns of caddisfly distribution in Canada and the U.S., results indicate that some caddisfly families with a larger body size are more diverse at northern latitudes, while smaller-bodied families show an opposite trend. Finally, to study environmental context and species distribution combine to determine the caddisfly community response to urbanization, I conducted a community science project in 2021. Volunteers collected caddisflies at 141 river sites across Canada and the U.S. with varying levels of urbanization. This project will shed light on whether caddisfly communities in certain areas are better equipped to tolerate the effects of urbanization because of the regional context and historical distribution of diversity and traits.
David Murray-Stoker, PhD student, Johnson Lab
Title: Evolutionary ecology of plant-microbe interactions in urban environments
Abstract: Urbanization is a dominant driver of environmental change that frequently alters the ecological setting of ecosystems. Determining how urban-driven environmental change alters the ecological and evolutionary dynamics of species interactions is important for understanding the drivers of biodiversity and ecosystem change in urban environments. Using the mutualism between white clover (Trifolium repens) and its rhizobial symbiont (Rhizobium leguminosarum symbiovar trifolii), my thesis addresses one central question: how does urbanization affect the evolutionary ecology of plant-microbe interactions? To establish how urbanization affects the ecology of the mutualism, I sampled white clover and rhizobia along an urbanization gradient in the Greater Toronto Area, ON, CA. I found that urban plants had reduced investment in the mutualism and acquired nitrogen (N) through fixation except at the urban and rural limits of the gradient, and soil N content was identified as the nexus linking urbanization to the mutualism. To determine if these ecological changes to the mutualism had evolutionary consequences, I conducted a large, common garden experiment to test for local adaptation between white clover and rhizobia. I found a spatial mosaic in local adaptation, with stronger local adaptation for rhizobia than white clover. While N addition strongly influenced plant biomass and nodule density, it did not consistently mediate patterns of local adaptation. The strength of local adaptation was positively related between white clover and rhizobia under N addition, but local adaptation was not directly influenced by urbanization. Together, my results demonstrate that urbanization alters the ecology and evolution of a legume-rhizobium mutualism.
Host: Prof. Emeritus Darryl Gwynne
https://utoronto.zoom.us/j/84984673393
Functional Genomics, Transcriptomics and Metabolomics: A Toolbox for Anticancer Drug Discovery.
Summary:
Background: MYC-amplified group 3 (G3) medulloblastoma (MB) represents a clinically aggressive subgroup of MB that is defined by leptomeningeal metastases, disease recurrence and particularly poor survivorship in comparison to other MB subgroups. There is an urgent need for a more thorough molecular understanding of G3 MB to develop more effective therapeutic modalities that will improve the durability of remission.
Methods: We mapped the functional G3 MB metabologenomic landscape via parallel genome-wide CRISPR-Cas9 screening and untargeted liquid chromatography-mass spectrometry (LC-MS) metabolomics. The tractability of potential therapeutic targets was assessed using patient-derived orthotopic preclinical models of G3 MB. Transcriptomics, functional genomics, and metabolomics were employed to discern a mechanism whereby inhibition of DHODH (dihydroorotate dehydrogenase), which is a mitochondrial-bound enzyme that facilitates de novo pyrimidine biosynthesis, selectively impairs G3 MB tumor cell fitness.
Results: Untargeted metabolomic profiling shows enrichment of de novo pyrimidine biosynthesis and depletion of salvage pyrimidine intermediates in G3 MB tumor cells in comparison to neural stem cells (NSC), which are the prospective cell-of-origin of MB. In agreement with reprogrammed nucleotide metabolism, DHODH was identified as a corresponding ‘druggable hit’ in the genetic screen. We demonstrate that genetic or pharmacological inhibition of DHODH selectively targets G3 MB brain tumor initiating cells (BTIC) while sparing normal NSC. We further show that MYC-amplified G3 MB tumors harbor subgroup-specific transcriptomic signatures that delineate enrichment of de novo pyrimidine biosynthesis, which sustains c-Myc activity via a metabolite-protein interaction.
Significance: Despite clear evidence favoring an altered metabolic landscape in MYC-driven cancers, there have been few reports into the role of metabolic reprogramming in MYC-amplified G3 MB. This metabolic vulnerability is hence not exclusive to MB and has recently been identified in other primary brain malignancies; current efforts are focused on establishing relevant biomarkers that will facilitate clinical translation.
https://utoronto.zoom.us/j/84984673393
Biomedical Communications: Exploring the Visual Communication of Science and Medicine
Description:
The Biomedical Communications Program has existed at the University of Toronto as an academic unit in one form or another since 1945. It now stands as the professional Master of Science in Biomedical Communications (MScBMC) Program, unique in Canada, and one of only a handful of similar graduate programs in North America. Although much has changed since its inception, the faculty and students still engage with all aspects of the visual communication of science and medicine. Over a two-year curriculum, the MScBMC Program engages students in the creation and evaluation of a range of visual tools, including medical illustration, media and user experience design, animation, and virtual simulations.
Our graduates are individuals with scientific training and the conceptual and practical skills necessary to design accurate and effective scientific media for a variety of audiences. Using examples of recent graduate student work, we will discuss how clear and compelling illustrations, animations, and simulations are vital to discovery and communication in science, medicine, and health.
Host: Shannon McCauley
Ephemeral Wetlands and Cryptic Salamanders: Conserving the Unknown
Ephemeral wetlands support unique species of conservation concern, but both the wetlands and the species in them are often overlooked and hard to find. Among the strangest of these are unisexual salamanders – an ancient lineage of females that borrow genes from five other species and dominate ponds throughout eastern North America. Protecting these systems depends on understanding processes that play out across various scales – from meiosis to metapopulation dynamics and land use history – but conservation is hampered by our poor understanding of the basic ecology. As scientists and as citizens, investing in core field natural history skills may be among the most important things we can do.
zoom link: https://utoronto.zoom.us/j/84984673393
Host: Marcus Dillon
Title: Evolution of Harpellales Fungi – Cryptic Gut Dwellers in Aquatic Insects
Abstract:
Harpellales is one of the most species-rich orders among early diverging fungi, encompassing 38 genera with over 200 species. These fungal species are obligate symbionts within insect guts and have been documented worldwide since their initial discovery in 1929. With the aid of comparative genomics, we have unraveled intriguing evolutionary signatures within the genome sequences of these gut-dwelling fungi. Our studies have led to the identification of a mosquito-originated polyubiquitin gene embedded in the genome of Zancudomyces culisetae (Harpellales), offering a unique insight into the horizontal gene transfers among eukaryotes. Furthermore, patterns of whole-genome duplications were discovered within certain lineages of Harpellales. One particularly interesting discovery involves the dual function of the stop codon, UGA, in Harpellales. This feature allows these fungi to synthesize Selenocysteine, which makes Harpellales distinctive in Kingdom Fungi. However, the draft quality of Harpellales genomes has posed challenges for in-depth investigations into these observations. To address this, our laboratory has recently assembled several reference-quality genomes of Harpellales. In this seminar, I will share the latest research advances in Harpellales, highlighting how these findings provide fresh insights into insect-fungus symbioses and shed light on the unconventional evolution of fungi in extreme environments, such as insect guts.
https://utoronto.zoom.us/j/84984673393
Host: Rosalind Murray
Vectors in Paradise: how mosquitoes interact with hosts across habitats in Puerto Rico, USA
There are over 3,600 species of mosquitoes in the world, however up until recently we did not know how many are responsible for transmitting pathogens that cause disease in humans. I will explore how we went about determining the exact number of medically relevant species, and how resilient pathogens are to the potential extermination of individual mosquito species. From there I plan to talk about my lab’s work on the island of Puerto Rico, USA, wherein we are investigating how vector communities are affected by socioeconomic factors and large hurricanes. Finally, I plan to discuss a new mosquito-pathogen-host system in the rain forests on the island, which has presented some intriguing mysteries for us to solve.
https://utoronto.zoom.us/j/84984673393
Host: Marc Johnson
Adventures in genetic toxicology: the quest to discover human germ cell mutagens
Genetic toxicologists are charged with characterizing and quantifying the mutagenic effects of environmental exposures to protect health. Although the foundation of genetic toxicology was rooted in understanding heritable genetic effects, after decades of research we lack consensus on whether a human germ cell mutagen even exists. This field of research was effectively derailed by the discovery of the causal relationship between somatic mutations and cancer, and the launch of science’s 'war on cancer'. Consequently, cancer research took precedence in the realm of genotoxicity testing. However, germ cell mutations lead to devasting genetic disorders and can have serious consequences at the population level. Indeed, we have quantified global socioeconomic repercussions stemming from even minor increments in germ cell mutation rates in a subset of disease-associated genes. This presentation will touch on highlights of three decades of work within my research program to unravel the connections between our environment and male germ cell mutagenesis, and understand its implications to humans. Starting with studies using unstable genetic elements (tandem repeat DNA) and culminating in the genomics revolution, I will describe our evolving understanding of the causes and landscape of germ cell mutagenesis. Our progress has been bolstered by the development of sophisticated error-corrected next-generation sequencing technologies, which are poised to dramatically transform this field. Although our germ cell are highly protected from mutagenesis, our research strongly suggests that chemical and physical agents associated with particulate air pollutants pose a risk to the male germ cell genome. It is clearly time to harness our technologies, synergize our efforts, and once and for all determine the environmental and social factors that drive germ cell mutagenesis and population health.
https://utoronto.zoom.us/j/84984673393
Host: Christoph Richter
Title: Zoogeochemistry: understanding animal roles in ecosystem nutrient cycles
Abstract: Animals are classically viewed as responders to ecosystems dynamics, shaped by the availability of habitat and resources in their environment. However, animals are increasingly understood to play critical roles in the functioning of ecosystems, particularly in nutrient cycles, which underpin many ecosystem processes. But where do these roles matter? How does animal diversity shape these roles? And how can we leverage "zoogeochemistry" to support ecosystem restoration?
https://utoronto.zoom.us/j/84984673393
Host: Marc Johnson
Urban evolution from phenotype to genotype in Anolis lizards
In the Anthropocene, life must rapidly evolve to survive unprecedented environmental challenges of an increasingly urbanized planet. In cities, where nearly every aspect is altered from the natural state, novel eco-evolutionary dynamics provide opportunities to answer fundamental questions in biology. One such question is to what extent is evolution shaped by random chance versus predictable processes? The retrospective nature of evolutionary study often means we must attempt to answer this question with incomplete evidence of the past pieced together in the present. However, leveraging the “natural experiment” of urbanization enables contemporary empirical study of evolution as it unfolds rapidly and repeatedly. Focusing on the evolutionary model system of Anolis lizards, I take an integrative approach to understanding adaptive phenotypic responses to urbanization. My work has demonstrated repeated phenotypic shifts in directions predicted by environmental divergence and habitat use and that confer a functional benefit to urban lizards. Exploring these differences at the genomic level, I found that phenotypic parallelism is underlain by genomic parallelism suggesting that urban lizards repeatedly arrive at the same adaptive solutions via the same genetic mechanisms. Together these studies paint a picture of a remarkably adaptable species of lizard that thrives in urban environments, providing insight into how evolution may shape life in an increasingly urbanizing world.
https://utoronto.zoom.us/j/84984673393
Protein Architecture of a Eukaryotic Genome
The goal of our research is to understand how genes are regulated. How do cellular and environmental signals toggle transcription regulatory proteins to be on or off, and by what mechanism do they deliver RNA polymerase to genes? To this end, one research goal is to map the protein-DNA architecture of whole genomes at single bp resolution. To achieve this, we developed ChIP-exo, which is ChIP-seq at ultra-high resolution. Using this assay, I will describe architectural themes of specific proteins at promoters and enhancers. These include sequence-specific transcription factors and their interactions with nucleosomes and transcription regulators like SAGA, SWI/SNF, and Mediator. These factors augment assembly of RNA polymerase II and the general transcription machinery into a transcription pre-initiation complex (PIC). One clear theme is that the architecture of inducible PICs is distinct from constitutive PICs.
Host: Ho-Sung Rhee
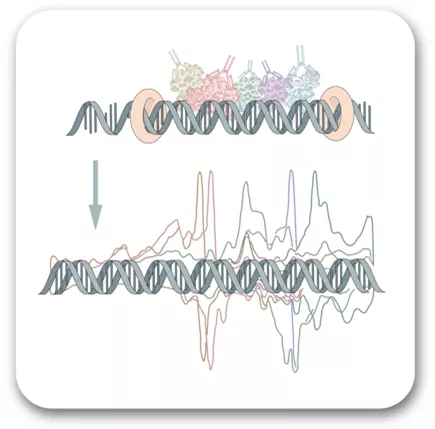
https://utoronto.zoom.us/j/84984673393
Host: Ingo Ensminger
Ecophysiology of Antarctic Plants: Freezing Tolerance vs Accelerated Regional Warming
León A. Bravo
Laboratory of Plant Physiology and Molecular Biology
Department Agronomical Sciences and Natural Resources
Universidad de La Frontera, Temuco Chile.
ABSTRACT
Antarctic vascular plants are freezing resistant. Deschampsia antarctica (Da) has LT50 of -27°C and Colobanthus quitensis (Cq) -15°C. This difference between both species is explained by several biochemical mechanisms that are only present in Da. For instance, the presence of apoplastic antifreeze proteins, accumulation of dehydrin-like proteins in vascular and epidermal tissues and sugar accumulation upon cold acclimation. But now this contrast with the accelerated warming (3.5°C in the last century) and heat waves experienced by Maritime Antarctica, especially during summer. This could cause plant cold de-acclimation making them vulnerable to stochastic frosts during the growing season. I will describe and discuss the effect of experimental in situ and laboratory warming on freezing resistance in Da and Cq. The results of in situ warming experiments using open top chambers (OTC) installed near Arctowski Station (King Gorge Island) showed that after two seasons of OTC exposure, LT50 increased slightly, 2°C in Cq, at two studied sites and 2.8°C at one of the 3 studied sites for Da. A contrasting situation was observed after four growing seasons in OTCs, while Cq showed a 2°C lower LT50 in plants grown in the OTC than untreated plants, Da showed no significant differences between treatments. Therefore, this suggest that warming 3 to 4°C above average temperature would not make these plants significantly more vulnerable to freezing. This was consistent with similar antifreeze activity observed in leaf apoplastic extracts of Da grown in OTC and in open areas. These results could be associated with the fact that OTC being a passive heating system does not exert night warming. Laboratory studies in which these two species were subjected to simulated day and night warming confirm this hypothesis. Preliminary results from nocturnal in situ warming systems will be presented and discussed.
https://utoronto.zoom.us/j/84984673393
EEB SuperSpeaker
Title: Large scale ecological responses to climate: carbon uptake and storage on land.
Abstract: Earth’s climate is changing with huge impacts for society and for ecosystems. Plants mediate the exchange of carbon, water and energy between the land and the atmosphere and so their changing response to climate also influences the future climate. In this talk, I will use a combination of in-situ observations, remote sensing and land surface models to investigate the controls on the land carbon sink and its persistence.
https://utoronto.zoom.us/j/84984673393
“Animating Molecular Machines”
My career has focused on creating visually rich animations depicting various molecular and cellular processes. During this presentation, I'll talk about my career path, and how I've found that 3D visualization is beneficial not only for communicating our ideas to broad audiences, but also for understanding and exploring the dynamics of complex molecular machines. I'll also discuss some of the challenges and future directions for modeling and visualizing dynamic molecular processes.
BMC Department Speaker
https://utoronto.zoom.us/j/84984673393
Xialolin Chou
A midbrain inhibitory pathway mediates the visual contextual modulation in the superior colliculus
As a fundamental attribute of sensory processing, our perception of objects is not only determined by the sensory inputs encoding the objects, but also influenced by their contexts. One common visual context is background motion, which occurs when our heads or the environment move. Many studies have shown that background motion has a profound influence on the perception of objects of interest, and related neural activity. However, it remains unclear how this contextual motion is represented in the brain and how it is employed to modulate visual processing. To address these questions, we used the circuit of the superior colliculus (SC), which is a prominent center of visual processing and integration in the midbrain. We found that background motion suppressed SC activity evoked by flashing objects, a novel model of visual contextual modulation. Moreover, we found that SC neurons received a remarkable amount of inhibition from the midbrain nucleus of optic tract (NOT), which is known to respond preferentially to background motion. To further study the functional role of the NOT-SC inhibitory pathway, we combined methodologies including electrophysiological recordings, circuit perturbations and behavioral assays, and demonstrated that this midbrain projection indeed encodes background motion signal and contributes to the modulation of SC activity and related visual guided behaviors by background motion. In conclusion, we identified a novel inhibitory midbrain pathway from NOT-DTN to SC which underlies the modulation of visual processing and perception by the contextual motion.
Xiaolin Chou is a postdoc fellow in Dr. Baohua Liu’s lab. He got his PhD in neuroscience in University of Southern California in 2020, studying the neural circuits for defensive behaviors. He then joined Dr. Xiaoke Chen’s lab in Stanford as a postdoc to study the thalamic circuits and contributions in associative learning. In September 2021, he moved to UTM and joined Dr. Baohua Liu’s lab. His research now focused on the investigation of functional roles of inhibition in visual processing and related behaviors.
Erik Etzler
Urban Ecology and Auditory Neuroethology in field crickets
Anthropogenic noise negatively affects some animal groups more so than others. Females crickets (Orthoptera: Gryllidae) reared in traffic noise have been reported to be both faster and slower to locate male song than females reared in silence. None of the studies tested whether differences were due to hearing or to decision-making. We reared female Teleogryllus oceanicus in traffic noise and silence, and had adults locate speakers playing male song amidst traffic noise or silence. We recorded the activity of the first-order auditory interneuron (AN2) in a subset of these individuals under identical acoustic conditions. Regardless of rearing treatment, crickets were slower to leave their shelter when presented with silence than with traffic noise. Crickets were also slower to leave their shelter if they were reared in traffic noise versus silence. At rest, crickets reared in traffic noise had higher baseline AN2 activity than those reared in silence, but rearing condition did not affect AN2 activity when male song was included. Our results demonstrate auditory and behavioural effects of long-term exposure to anthropogenic noise. Further, they support the idea that silence itself is an unnatural and potentially adversive acoustic condition for many animals.
Species interactions at tundra transitions, where threatened predators create hotspots of productivity and diversity
Jim Roth, University of Manitoba
https://utoronto.zoom.us/j/84984673393
Abstract: Cyclic predator-prey dynamics, as seen in Arctic foxes and lemmings, illustrate the strong link between predators and their prey, and may dramatically impact many other species. We examined these direct and indirect interactions among species at the taiga-tundra transition zone in northern Manitoba, where the impacts of climate change are particularly strong. Changing snow and ice conditions, and the simultaneous encroachment of southern species onto the tundra, can substantially alter these species interactions. Yet the hotspots of productivity created by foxes fertilizing den sites, which stimulates plant growth and attracts wildlife, persistently maintain landscape heterogeneity, enhancing biodiversity.
Bio:
Jim Roth is a Professor in Biological Sciences at the University of Manitoba.
My research focuses on species interactions and population dynamics, with an emphasis on Arctic environments. I’m interested in how prey availability affects predators and their alternative prey, particularly within food webs that cross ecosystem boundaries (marine to terrestrial) and novel interactions involving species that have expanded their range as the environment changes. I’m also interested in predators that engineer the Arctic ecosystem by concentrating nutrients, and the cascading impact on a diversity of species. I first started working on Arctic food web interactions during my PhD at the University of Minnesota, which I completed in 1998. I continued to work on boreal species interactions as a post doc at the University of Idaho, and subsequently as a Research Assistant Professor at the University of Central Florida. Since moving to Winnipeg in 2009, introducing students to northern research has been a priority. I have taken over 100 university students to conduct research at the taiga-tundra transition zone in northern Manitoba, and collect data for my long-term research program examining northern biodiversity.
https://www.churchillfoxproject.org/
Host: Peter Kotanen
Host: Steven Short
Beautiful Shades of Brown – Illuminating Photosynthesis in the Ocean
Photosynthesis in plants and algae teeters on the knife edge of necessity and catastrophe. They need to be able to collect enough light energy to drive the primary reactions of photosynthesis. But absorbing too much light leads to oxidative stress, cellular damage and death. Decades of research have revealed how vascular plants toggle light collection to balance photosynthesis and photo-protection. However, algae appear to have come up with independent solutions to the same problem. I will talk about our systems-level observations of algal biology to discover the unexpected biochemical pathways that have evolved to allow brown-colored algae to use green light for photosynthesis. I will then discuss a “rheostat” that permits these same organisms to flow between a light harvesting state and light energy dissipation state. Over the course of the seminar, I will relate these findings back to the ecophysiology of algae in the aquatic environment and how these processes are important for their ecological success in the oceans.
https://utoronto.zoom.us/j/84984673393
CRISPR-based functional genomic tools and applications to understanding antifungal resistance and fungal virulence
Abstract:
Opportunistic Candida pathogens are a leading cause of fungal infections, and new functional genomic tools enable our ability to better study the biology of these important pathogens. Here, we describe the development and optimization of CRISPR-based tools for functional genomic analysis in Candida albicans. We have developed two powerful technologies: CRISPR interference (CRISPRi) and CRISPR activation (CRISPRa), for applications in Candida species. We demonstrate the ability of these systems to robustly repress or induce gene expression in C. albicans, including in drug-resistant clinical isolates, and further demonstrate the application of these systems to characterize factors involved in resistance to antifungal drugs and fungal virulence. Since, unlike classic CRISPR systems, these platforms do not require a DNA repair construct, the simplicity of this system lends itself to high-throughput strain generation. Using a highly efficient, high-throughput cloning strategy, we are able to efficiently and rapidly generate large numbers of fungal mutant strains that over- or under-express any gene of interest, providing a powerful new tool for functional genomic analyses in fungal pathogens. We are applying these CRISPR-based functional genomic libraries to study antifungal drug resistance on a large scale, and understand mechanisms of fungal-host colonization and virulence.
Host: Alex Nguyen Ba
https://utoronto.zoom.us/j/84984673393
Anna Metaxas, Department of Oceanography, Dalhousie University
Connectivity is essential to meet conservation targets for the global ocean
The importance of ecological connectivity is being recognized in most major international fora dealing with biodiversity conservation, such as the Global Biodiversity Framework, the agreement on Biodiversity Beyond National Jurisdiction, and the regional environmental management plans for deep-sea mining by the International Seabed Authority. However, this recognition is currently not well-coupled with implementation in the oceans. I will present the current state of the incorporation of marine ecological connectivity in global conservation targets, and address some of the gaps and limitations for moving forward. Using some of our own research, I will attempt to provide some ideas on future directions.
Host: Cassidy D'Aloia
https://utoronto.zoom.us/j/84984673393
Host: Mark Currie
Asymmetric Histone Inheritance: Establishment, Recognition, Function, and Conservation
We found that during asymmetric division of stem cells, preexisting old histones are selectively retained in the stem cell, whereas newly synthesized histones are enriched in the daughter cell that is committed to differentiation. This process provides an important mechanism that allows the two daughter cells to each inherit different epigenetic information from a single cell division. These intriguing findings urge us to better understand how cells maintain their epigenetic memories or reset their epigenome. We found that the mechanism and readout of asymmetric histone inheritance involve at least three-steps, wherein old and new histones are asymmetrically incorporated during DNA replication (Step 1) and are segregated in a biased manner during mitosis (Step 2), driving differential inheritance of key factors to regulate distinct cellular events in the resulting daughter cells (Step 3). We have developed new tools and methods to explore the molecular and cellular mechanisms. Recently, we studied this phenomenon in multiple systems across species and identified conserved mode of histone inheritance. I will discuss the updated information regarding the biological significance and generality of asymmetric histone inheritance in multicellular organisms.
https://utoronto.zoom.us/j/84984673393
Extracting meaning from high-throughput functional genomics data with Bayesian statistics
High-throughput sequencing-based functional genomics approaches have transformed molecular infection biology into an increasingly data-driven science. However, extracting meaning from complicated experimental designs remains a challenge. Bayesian statistics offers a promising methodology to capture biological and technical parameters of interest from complex experiments by allowing the analyst to directly model and test hypotheses about the data generating process. In this talk, I will show how a principled Bayesian modeling approach to investigating RNA decay dynamics has led us to substantially revise global estimates of RNA half-life in Salmonella enterica and enables us to perform transcriptome-wide differential stability analyses to investigate the regulatory impact of bacterial RNA-binding proteins. I will further show how a similar modeling approach has allowed us to design and implement a saturating genome-wide transposon screens for Shigella flexneri host cell invasion factors in a realistic human organoid model of intestinal infection while accounting for important experimental factors like bottleneck effects. In sum, our Bayesian modeling approach provides a principled methodology for the analysis and interpretation of complex functional genomics experiments.
Host: Marcus Dillon
Host: Baohua Liu
Unraveling lateral habenula neuronal outputs in normal and depressive behaviors.
The lateral habenula (LHb) receives signals from the basal ganglia involved in the selection of actions, and from various nuclei of the limbic system that play roles in the processing of various cognitive and affective signals. In turn, the LHb sends excitatory efferents to the dopaminergic ventral tegmental area (VTA), the serotoninergic dorsal raphe nucleus (DRN), and the GABAergic rostromedial tegmental nucleus (RMTg), which inhibits dopaminergic centers. The LHb is therefore well positioned to integrate emotion into the selection of actions. The LHb has emerged as a pivotal brain region implicated in depression, displaying hyperactivity in human and animal models. In my lab, we combine fiber photometry calcium imaging and and optogenetics approaches in freely behaving mice, viral tracing approaches and whole-cell recordings to unravel the functions of the LHb neuronal outputs in signal processing and behavioral control in normal and in animal models of depression.
Special Topic Speaker
https://utoronto.zoom.us/j/84984673393
ARTIST TALK by Diane Borsato
Diane Borsato will present an artist talk about recent environmentally-themed projects she has made together with mushroomers, beekeepers, orchardists and other artists and naturalists in her multidisciplinary contemporary art practice.
Diane Borsato is an award-winning artist, writer, and educator - who has worked closely with other artists and amateur naturalists, including co-ordinating a day-long foraging and stargazing event for amateur mushroomers and astronomers (Terrestrial/Celestial, 2010), creating a performance of silence and stillness with 100 beekeepers (Your Temper, My Weather, 2013), and planting a rare community apple orchard as public art (ORCHARD, 2019 - ongoing) along with many other multidisciplinary projects.
She has performed and exhibited across Canada at venues including the Art Gallery of Ontario, The Power Plant, the AGYU, the Art Museum at the University of Toronto, the Walter Philips Gallery at the Banff Centre for the Arts, the Esker Foundation, the Vancouver Art Gallery, the National Art Centre, Fogo Island Arts and at the Toronto Biennial of Art; and internationally at galleries and museums in the US, Mexico, Australia, France, Germany, Iceland and Japan among others. She recently published two books, Outdoor School: Contemporary Environmental Art (2021) co-edited with Amish Morrell, and MUSHROOMING: The Joy of the Quiet Hunt (2022) – a guide to mushroom foraging and an exploration of fungi in contemporary art. She is also an Associate Professor of Experimental Studio at the University of Guelph, where she teaches courses exploring conceptual art, video and performance art; along with social, site-responsive and environmental art practices. See her work at www.dianeborsato.net
“ Establishing an analytical base to support the exposome disease atlas”
Dr. Walker is CSB’ Super Speaker. Dr. Walker will present via zoom: https://utoronto.zoom.us/j/84984673393
Everyone is invited to attend either via zoom or in person, IB335
Over a lifetime, humans experience thousands of chemical exposures that may contribute to increased risk of neurodegenerative diseases. A more complete estimate of environmental exposures across the lifespan and how these may contribute to disease outcomes would be a transformative research initiative. The use of high-resolution, mass spectrometry (HRMS) provides a key platform for assessing the exposome and provides measures of thousands of chemical signals in a single human sample; however, application of internal exposome profiling in large populations has been limited due to challenges in sample throughput, instrument robustness, and data handling methods. To support large-scale population research for exposome epidemiology, we have established a high-throughput untargeted HRMS analytical framework combining parallel analysis by liquid (LC) and gas (GC) chromatography to enable robust analysis of up to 20,000 blood samples per year. Sample preparation is achieved using low-cost and open-source automated liquid handlers allowing parallel processing of extracts for LC and GC analysis from a single blood aliquot of 150µL, eliminating operator effects and allowing daily preparation of 96 samples in under 3.5 hours. Exposome profiles are measured using five different HRMS analytical configurations that include reverse phase and HILIC chromatography with both positive and negative ionization, and GC-HRMS to provide detection of 60,000-100,000 chemical signals. Resources for annotation include a standard library of over 7,000 compounds covering a wide range of environmental, drug and endogenous metabolites. The resulting exposome profiles are being assembled within a framework built upon SQL databases, providing a cumulative resource for assembling exposome-disease atlases and laying the foundation for the Human Exposome Project. Development of a robust and scalable analytical framework for large-scale exposome epidemiology to study human disease is a critical first step to provide a robust foundation for exposome research and to facilitate development of a knowledge base of environmental chemicals, their products, distributions, and associated effects.
Douglas Walker, PhD, is an Associate Professor in the Gangarosa Department of Environmental Health at Emory University, where he leads the Comprehensive Laboratory for Untargeted Exposome Science (CLUES). He is an environmental engineer and analytical chemist with training in metabolomics and developing EWAS (exposome-wide association study) methodologies for environmental health and precision medicine research. Dr. Walker’s research focuses on continued development and application of advanced analytical strategies for measuring the occurrence, distribution and magnitude of previously unidentified environmental exposures and assisting in delineating the mechanisms underlying environment-related diseases in humans. The approaches he developed show it is possible to measure over 100,000 chemical signals that include exposure biomarkers, nutrients, dietary chemicals and associated biological response in a high-throughput and cost-effective manner, establishing a foundation for operationalizing the exposome framework for precision medicine. Ongoing research projects are now focused on using high-throughput exposome methods to establish disease-exposome atlases, and development of methods for measuring biomarkers of complex exposures of emerging concern, including microplastics, e-waste and polyfluorinated chemicals.
Sep 15, 2023
Sep 29, 2023
Oct 6, 2023
Oct 20, 2023
Oct 27, 2023
Nov 3, 2023
Nov 10, 2023
Nov 17, 2023
Nov 24, 2023
Dec 1, 2023
Jan 19, 2024
Jan 26, 2024
Feb 2, 2024
Feb 16, 2024
Mar 1, 2024
Mar 8, 2024
Mar 15, 2024
Mar 22. 2024
Apr 5, 2024
