Arnaud N’Guessan' first 1st authored paper in eLife
We are thrilled to announce that Arnaud N’Guessan, PhD student, Alex Nguyen Ba Lab, published his first 1st authored paper: Refining the resolution of the yeast genotype-phenotype map using single-cell RNA-sequencing in eLife
https://doi.org/10.7554/eLife.93906.1
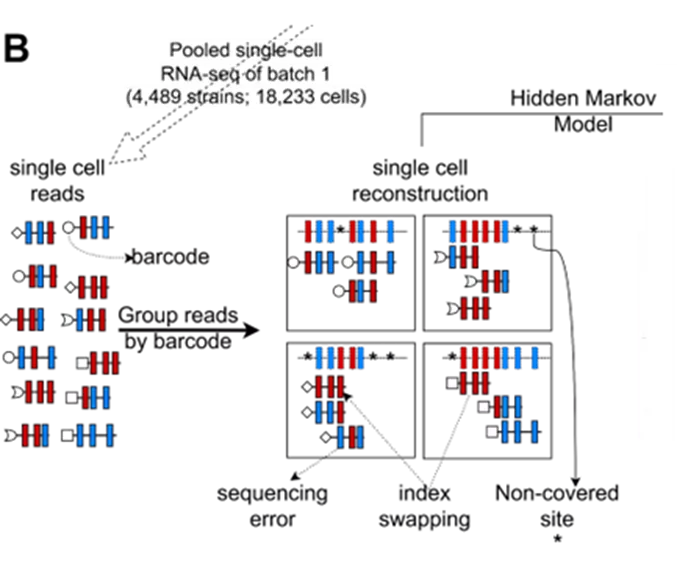
Genotype-phenotype mapping (GPM) or the association of trait variation to genetic variation has been a long-lasting problem in biology. The existing approaches to this problem allowed researchers to partially understand within- and between-species variation as well as the emergence or evolution of phenotypes. However, traditional GPM methods typically ignore the transcriptome or have low statistical power due to challenges related to dataset scale. Thus, it is not clear to what extent selection modulates transcriptomes and whether cis- or trans-regulatory elements are more important. To overcome these challenges, we leveraged the cost efficiency and scalability of single-cell RNA sequencing (scRNA-seq) by collecting data from 18,233 yeast cells from 4,489 segregants of a cross between the laboratory strain BY4741 and the vineyard strain RM11-1a. More precisely, we performed eQTL mapping with the scRNA-seq data to identify single-cell eQTL (sc-eQTL) and transcriptome variation patterns associated to fitness variation inferred from the segregants’ bulk fitness assay. Due to the larger scale of our dataset, we were able to recapitulate results from decades of work in GPM from yeast bulk assays while revealing new associations between phenotypic and transcriptomic variations. The multidimensionality of this dataset also allowed us to measure phenotype and expression heritability and partition the variance of cell fitness into genotype and expression components to highlight selective pressure at both levels. Altogether these results suggest that integrating large-scale scRNA-seq data into GPM improves our understanding of trait variation in the context of transcriptomic regulation.
Arnaud's bio
I am a 3rd year CSB PhD student in Dr. Alex Nguyen Ba's lab. In the lab, we use high-throughput technologies such as single-cell RNA-sequencing, and statistical models to answer biological questions in large-scale datasets. With these tools, I am interested in understanding how the evolution of the transcriptome shapes within- and between-species diversity, contributes to adaptation and could be leveraged for disease treatment and diagnosis.