Microbial Ecology and Population Dynamics
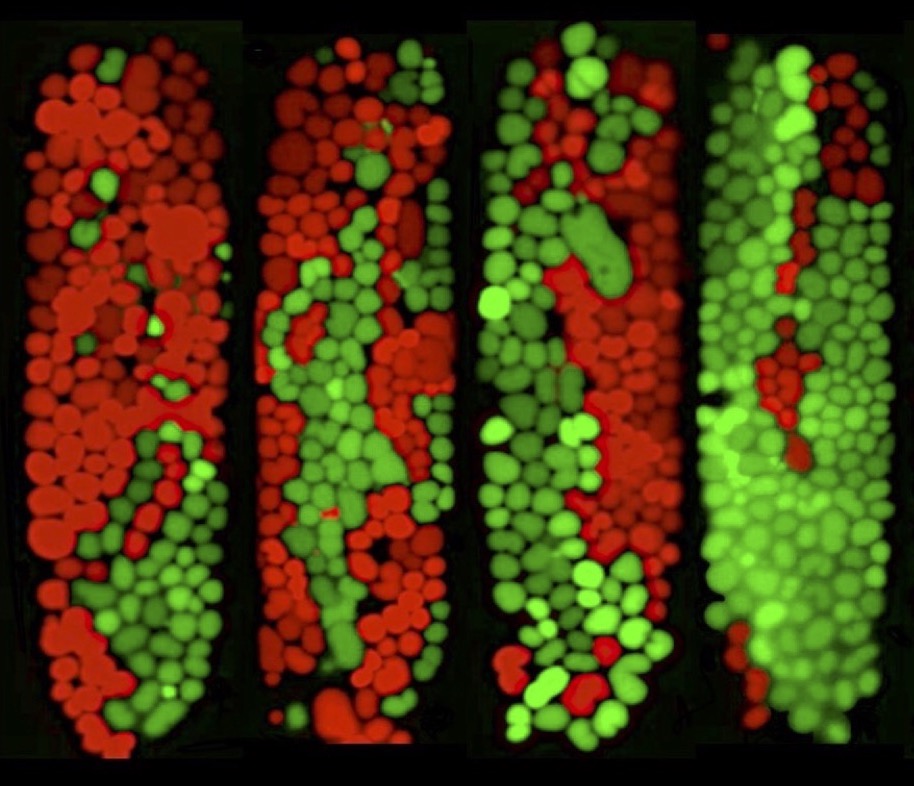
Bacteria primarily exist within dense and complex communities, like biofilms or the gut microbiome, where they compete for limited space and resources. These microbial consortia display heirarchical levels of spatial patterning much like eukaryotic cells within macro-organisms. This organization, resulting from the complex dynamics of competitive and cooperative interactions, can lead to emergent properties of functional significance to the consortia as a whole.
We are studying the single-cell population dynamics of small microbial systems where fluctuations and stochasticity dominate, to better understand the strategies employed by bacteria to cooperate or compete with other bacteria.
Mechanics limits ecological diversity and promotes heterogeneity in confined bacterial communities,
T. Ma, J. Rothschild, F. Halabeya, A. Zilman, and J. N. Milstein, Proceedings of the National Academy of Sciences 121 (20) e2322321121 (2024).
Spatial exclusion leads to "tug-of-war" ecological dynamics between competing species within microchannels,
J. Rothschild, T. Ma, J. N. Milstein and A. Zilman, PLoS Computational Biology 19(12): e1010868 (2023).
Single-Molecule Imaging and Optical 'Omics

Recent advances in optical imaging have been so dramatic that light microscopy can now be used to directly quantify protein or nucleic acid numbers, modifications and interactions. This has given rise to the new fields of ‘optical proteomics’ and ‘optical genomics’.
We are developing quantitative approaches to single-molecule microscopy that will help us to understand the organizational principles of cells by mapping out their internal structure and quantifying the intracellular abundance of proteins and nucleic acids. This information can uncover functional oligomerization by signalling receptors such as GPCRs or provide insight into stochastic varation among populations of bacteria.
Single-Molecule Counting Applied to the Study of GPCR Oligomerization,
J. N. Milstein, D. F. Nino, X. Zhou and C. Gradinaru , Biophys. J. 121, 3175-3187 (2022).
An Expectation-Maximization Approach to Quantifying Protein Stoichiometry with Single-Molecule Imaging,
A. Boonkird, D. F. Nino and J. N. Milstein, Bioinformatics Advances 1, vbab032 (2021).
Estimating the Dynamic Range of Quantitative Single-Molecule Localization Microscopy,
D. Nino and J. N. Milstein, Biophys. J. 120, 3901-3910 (2021).
Nanoscopic Stoichiometry and Single-Molecule Counting,
D. Nino, D. Djayakarsana and J. N. Milstein, Small Methods 3, 1900082, (2019).
Molecular Counting with Localization Microscopy: A Bayesian estimate based on fluorophore statistics,
D. Nino, N. Rafiei, Y. Wang, A. Zilman and J. N. Milstein, Biophys. J. 112, 1777-1785 (2017).
Uncovering the Biophysical Mechanisms Behind Gene Silencing
 Bacteria can rapidly adapt to a changing environment by acquiring genes from viruses or other bacteria. Expressing these genes, however, may entail a fitness cost putting the bacteria at a competitive disadvantage or, in the worst case, lead to cell death. The incorporation and eventual expression of foreign DNA is, therefore, carefully controlled. Newly acquired, foreign DNA must, at least initially, be recognized as such and silenced.
Bacteria can rapidly adapt to a changing environment by acquiring genes from viruses or other bacteria. Expressing these genes, however, may entail a fitness cost putting the bacteria at a competitive disadvantage or, in the worst case, lead to cell death. The incorporation and eventual expression of foreign DNA is, therefore, carefully controlled. Newly acquired, foreign DNA must, at least initially, be recognized as such and silenced.
The protein H-NS, found in common bacteria like E. coli and Salmonella, and Lsr2, found in the pharmaceutical factories Streptomyces, are examples of silencing proteins that target foreign DNA. These proteins are known to somehow interfere with genetic transcription. Bulk biochemical and genomic approaches to understanding gene silencing by H-NS and Lsr2 have been employed for decades. Only recently, with the advent of single-molecule measurements, have researchers begun to gain a fundamental understanding of the biophysical mechanisms through which H-NS and Lsr2, and associated co-regulatory proteins, function to regulate gene expression.
An Ultra-Stable and Dense Single-Molecule Click Platform for Sensing Protein-Deoxyribonucleic Acid Interactions,E. Visser, J. Miladinovic and J. N. Milstein, Small Methods 5, 2001180, (2021).
Growth Phase Dependent Chromosome Condensation and H-NS Protein Redistribution in E. coli Under Osmotic Stress,
N. Rafiei, M. Cordova, W. W. Navarre and J. N. Milstein, J. Bacteriol. 23, e00469-19 (2019).
Xenogeneic silencing and its impact on bacterial genomes,
K. Singh, J. N. Milstein and W. W. Navarre, Annu. Rev. Microbiol. 70, 199-213 (2016).
Optical Engineering and Force Spectroscopy

We are developing a next generation of axial optical tweezers that can manipulate small segments of DNA with an extremely high degree of spatial and temporal precision. These tools will enable us to explore novel regulatory mechanisms such as sequence dependent effects on protein-DNA interactions, which are inaccessible via current technology.
Our focus is on using light-shaping techniques, resulting in what are often called holographic optical tweezers, to correct for aberrations that would otherwise interfere with single-molecule measurements and to engineer the focal spot for novel applications of optical trapping.
Extending the Range of Rupture Force Measurements with Axial Optical Tweezers,Z. Zhang and J. N. Milstein (preprint @ bioRxiv, 2023 ).
Extending the Photobleaching Lifetime in the Presence of an Optical Trap by Wavefront Engineering,
Z. Zhang and J. N. Milstein, J. Optics 22, 095301 (2020).
Accounting for polarization in the calibration of a donut beam axial optical tweezers,
R. Pollari and J. N. Milstein PLoS ONE 13(2): e0193402 (2018).
Bioimage Analytics
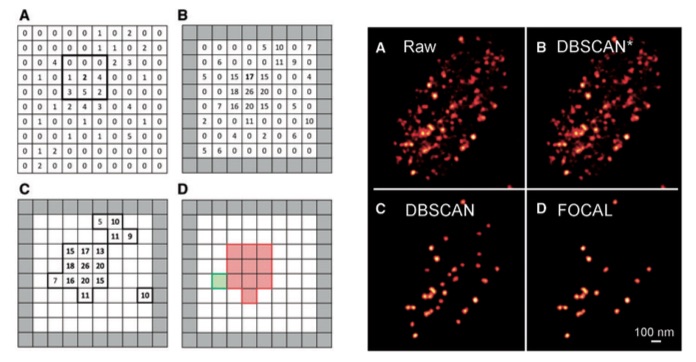 With the advent of single-molecule localization microscopy (SMLM), images of cellular structure and organization can be acquired with visible light at a spatial resolution well surpassing the diffraction limit. SMLM is increasingly being employed in cells to image and quantitatively analyze various protein complexes forming tens to hundreds of nm assemblies, from membrane receptors to nucleosome bundles.
With the advent of single-molecule localization microscopy (SMLM), images of cellular structure and organization can be acquired with visible light at a spatial resolution well surpassing the diffraction limit. SMLM is increasingly being employed in cells to image and quantitatively analyze various protein complexes forming tens to hundreds of nm assemblies, from membrane receptors to nucleosome bundles.
We are developing clustering algorithms for detecting and analyzing protein aggregates in SMLM datasets and have already applied the technique to study transcription factories within cell nuclei. The software package is currently made available on our lab website and, while it only works on 2-dimensional datasets, we will soon be releasing a more extensive version that directly clusters 3D SMLM data.
FOCAL3D: A 3-dimensional clustering package for single-molecule localization microscopy,D. Nino, D. Djayakarsana and J. N. Milstein, PLoS Computational Biology, 16(12): e1008479 (2020).
Fast Optimized Cluster Algorithm for Localizations (FOCAL): A spatial cluster analysis optimized for super-resolved microscopy,
A. Mazouchi and J. N. Milstein, Bioinformatics 32(5), 747–754 (2016).