
UTM student turns to 80s video game to combat cancer
A medicinal chemist at the U of T Mississauga is drawing on the popular classic video game Tetris to battle cancer.
Muhammad Murtaza Hassan, a PhD student in Professor Patrick Gunning’s lab, has turned to mainstream gameplay, harnessing the concept of fitting geometric patterns together to create an inhibitor that targets a specific protein associated with cancer.
“When medicinal chemists want to target cancer cells selectively, over healthy cells, they often target a distinguishing factor of a cancer cell over a healthy cell,” Hassan says, explaining there are certain proteins that promote the hallmarks of cancer, like uncontrolled proliferation. “What medicinal chemists do is target these proteins that are associated with these hallmarks of cancer.
Hassan wanted to target HDAC8, a protein involved in neuroblastoma, as well as some viruses.
He turned his attention to the protein while he was working with the larger group of HDAC proteins in general. He noticed some trends when he added chemical groups to the proteins, which prompted him to examine the shape of HDAC8 specifically.
“It’s kind of L-shaped,” he says.
Like a game of Tetris, Hassan started to design a molecule that would adopt a complementary shape to fit the protein.
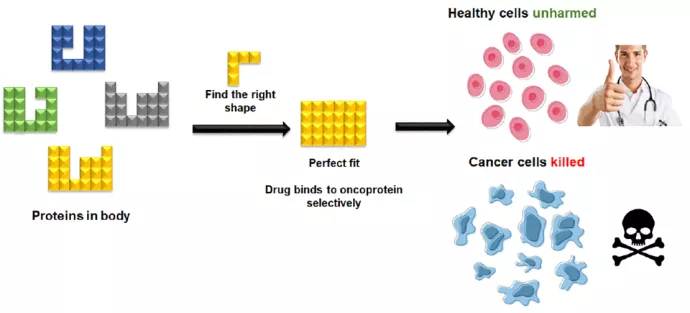
Molecules, of course, are far more complicated than the 1984 tile-matching game. Unlike Tetris, it’s not enough to simply have a complementary shape between the protein and drug. The chemical groups between the two must also match, and in the correct position.
“You can imagine there’s a large number of possible conformations,” Hassan says. “You have type of chemical group, position of chemical group and you have the orientation of the molecule.”
Sitting at the bottom of the L-shaped protein tunnel is zinc, so Hassan put a chemical group on the complementary-shaped molecule that “latches on” to that zinc. The specific chemical group and positioning allowed the molecule to bind to the protein while the shape made it target HDAC8 specifically.
“This type of approach made one of the most selective HDAC8 inhibitors known to date,” says Hassan, who has published the results of his study in the Journal of Medicinal Chemistry.
It took Hassan about a year to make the inhibitors, which have since been tested with the help of a pediatric oncologist in Germany who analyzed the molecules on neuroblastoma cancer cells. Hassan says the tests showed that the molecules were interacting with HDAC8 in the cancer cells.
Hassan, who completed his undergraduate studies at UTM in chemistry and math before continuing to York University for his master’s, says he originally thought chemistry was boring because he didn’t fully realize its potential. It wasn’t until he put on what he terms his “medicinal chemistry glasses,” that he started to think of it like a molecular puzzle. Now he likens chemistry to engineering at a molecular level, where the goal is curing diseases.
“Had I not studied under (Patrick Gunning), I may not have ever seen this,” Hassan says.
Using that analogy of a molecular puzzle and borrowing from a classic video game, Hassan has now found a rational approach to selectively focus on a specific protein associated with cancer and other illnesses. He notes that when it comes to targeting proteins, people often go in without knowing the exact shape of the molecule. “That means they’re basically playing Tetris blind a lot of the time,” Hassan says.
“This study has given us a very selective way of targeting a protein,” he continues, noting this additional information could be used in the future to treat cancer, viruses and maybe even aging.
Read more:
